Introducing KitikiPlot, a Python library designed for visualizing sequential and time-series categorical “Sliding Window” patterns. This progressive device is designed to empower information practitioners throughout varied fields, together with genomics, air high quality monitoring, and climate forecasting to uncover insights with enhanced readability and precision. Designed with simplicity and flexibility, it integrates seamlessly with Python’s information ecosystem whereas providing visually interesting outputs for sample recognition. Let’s discover its potential, and rework the best way of analyzing categorical sequences.
Studying Targets
- Perceive the KitikiPlot sliding window visualization method for sequential and time-series categorical information.
- Discover its parameters to tailor visualizations for particular datasets and purposes.
- Apply KitikiPlot throughout varied domains, together with genomics, climate evaluation, and air high quality monitoring.
- Develop proficiency in visualizing complicated information patterns utilizing Python and Matplotlib.
- Acknowledge the importance of visible readability in categorical information evaluation to boost decision-making processes.
This text was printed as part of the Knowledge Science Blogathon.
KitikiPlot: Simplify Complicated Knowledge Visualization
KitikiPlot is a strong visualization device designed to simplify complicated information evaluation, particularly for purposes like sliding window graphs and dynamic information illustration. It presents flexibility, vibrant visualizations, and seamless integration with Python, making it ideally suited for domains reminiscent of genomics, air high quality monitoring, and climate forecasting. With its customizable options, KitikiPlot transforms uncooked information into impactful visuals effortlessly.
- KitikiPlot is a Python library for visualizing sequential and time-series categorical “Sliding Window” information.
- The time period ‘kitiki‘(కిటికీ) means ‘window‘ in Telugu.
Key Options
- Sliding Window: The visible illustration consists of a number of rectangular bars, every akin to information from a selected sliding window.
- Body: Every bar is split into a number of rectangular cells referred to as “Frames.” These frames are organized side-by-side, every representing a worth from the sequential categorical information.
- Customization Choices: Customers can customise home windows extensively, together with choices for colour maps, hatching patterns, and alignments.
- Versatile Labeling: The library permits customers to regulate labels, titles, ticks, and legends based on their preferences.
Getting Began: Your First Steps with KitikiPlot
Dive into the world of KitikiPlot with this quick-start information. From set up to your first visualization, we’ll stroll you thru each step to make your information shine.
Set up KitikiPlot utilizing pip
pip set up kitikiplotImport “kitikiplot”
import pandas as pd
from kitikiplot import KitikiPlotLoad the dataframe
Thought of the information body ‘weatherHistory.csv’ from https://www.kaggle.com/datasets/muthuj7/weather-dataset.
df= pd.read_csv( PATH_TO_CSV_FILE )
print("Form: ", df.form)
df= df.iloc[45:65, :]
print("Form: ", df.form)
df.head(3)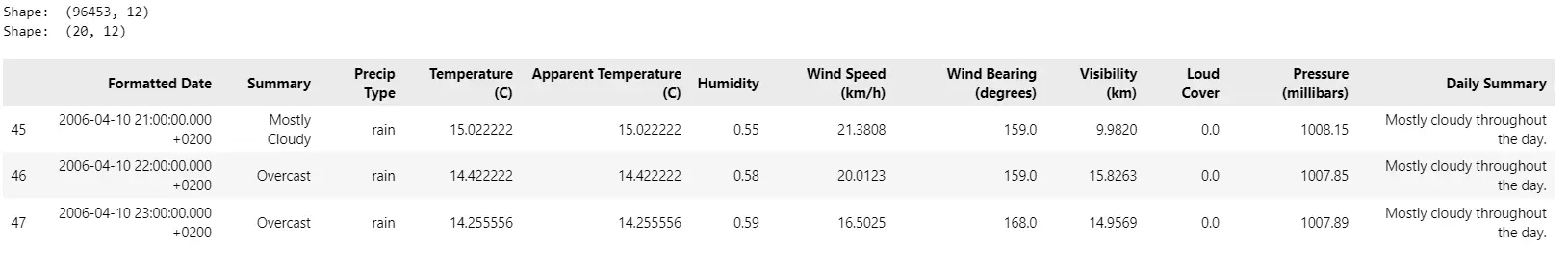
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( ) 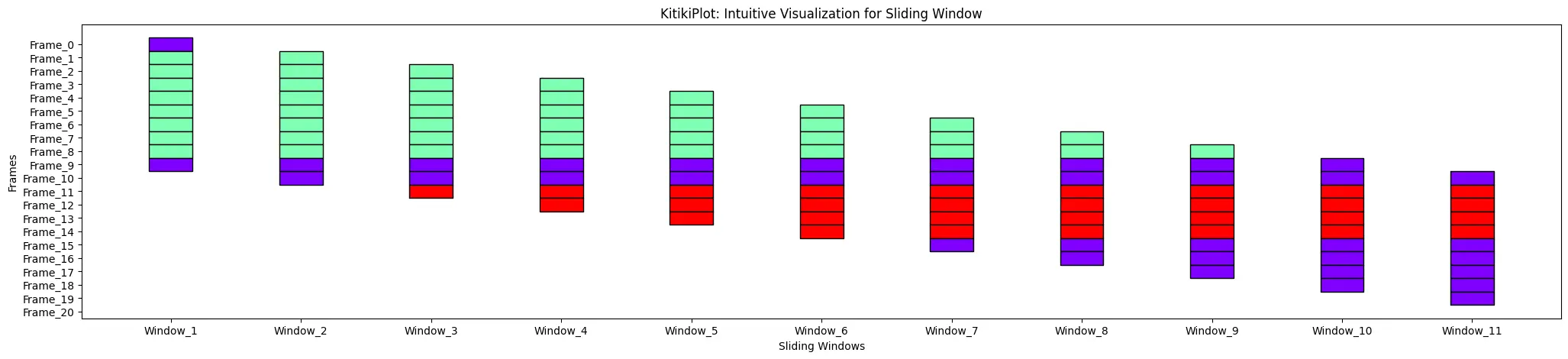
Understanding KitikiPlot Parameters
To completely leverage the ability of KitikiPlot, it’s important to know the varied parameters that management how your information is visualized. These parameters can help you customise points reminiscent of window dimension, step intervals, and different settings, guaranteeing your visualizations are tailor-made to your particular wants. On this part, we’ll break down key parameters like stride and window_length that will help you fine-tune your plots for optimum outcomes.
stride : int (optionally available)
- The variety of components to maneuver the window after every iteration when changing a listing to a DataFrame.
- Default is 1.
index= 0
ktk= KitikiPlot( information= df["Summary"].values.tolist(), stride= 2 )
ktk.plot( cell_width= 2, transpose= True )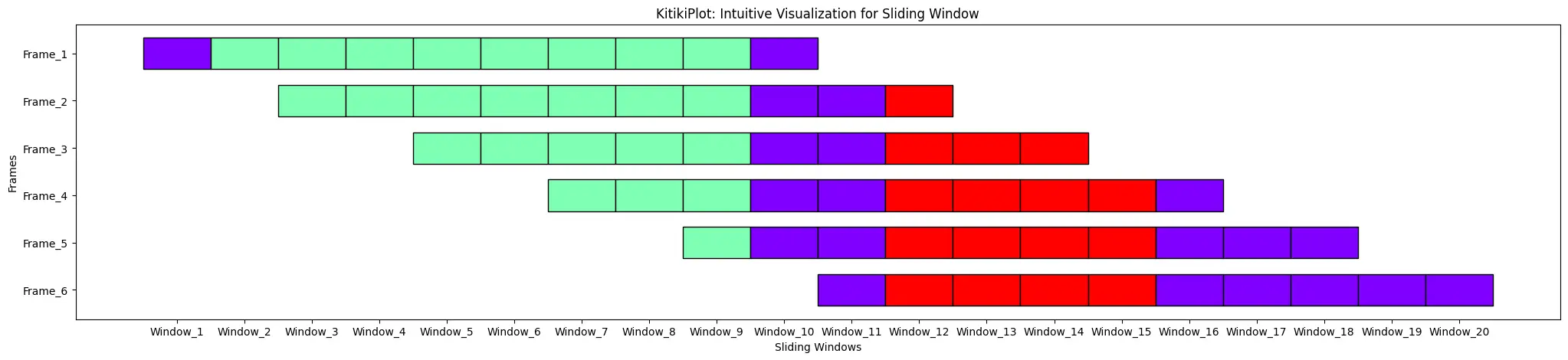
window_length : int (optionally available)
- The size of every window when changing a listing to a DataFrame.
- Default is 10.
index= 0
ktk= KitikiPlot( information= df["Summary"].values.tolist(), window_length= 5 )
ktk.plot( transpose= True,
xtick_prefix= "Body",
ytick_prefix= "Window",
cell_width= 2 ) 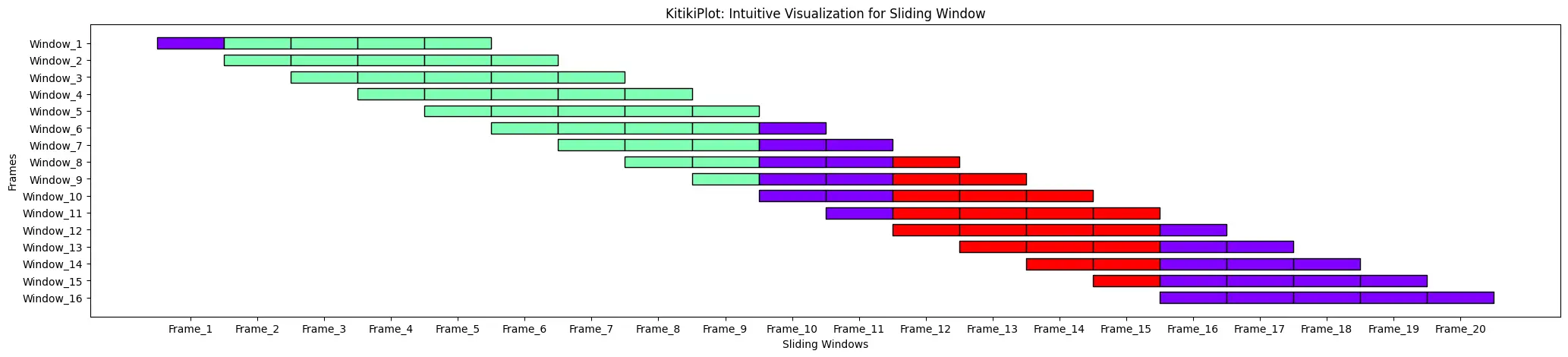
figsize : tuple (optionally available)
- The dimensions of the determine (width, top).
- Default is (25, 5).
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( figsize= (20, 8) )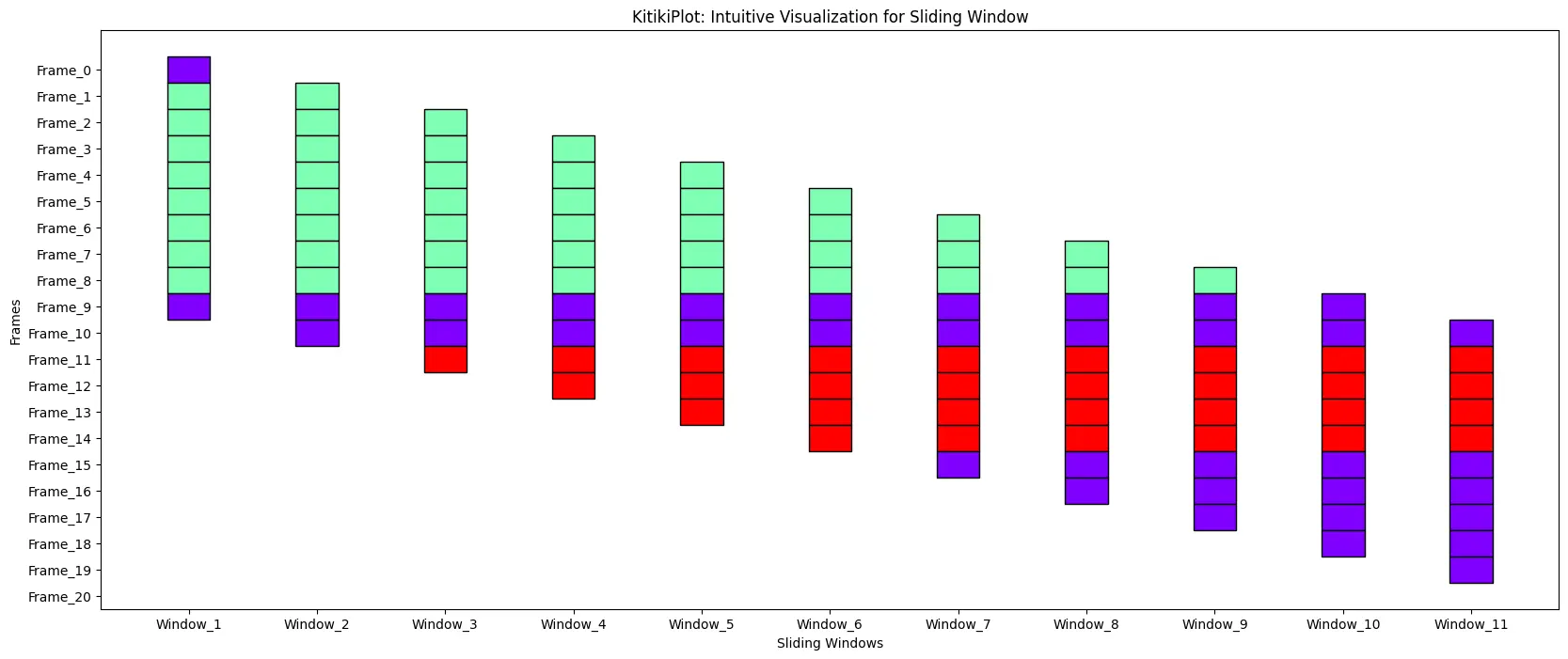
cell_width : float
- The width of every cell within the grid.
- Default is 0.5.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( cell_width= 2 )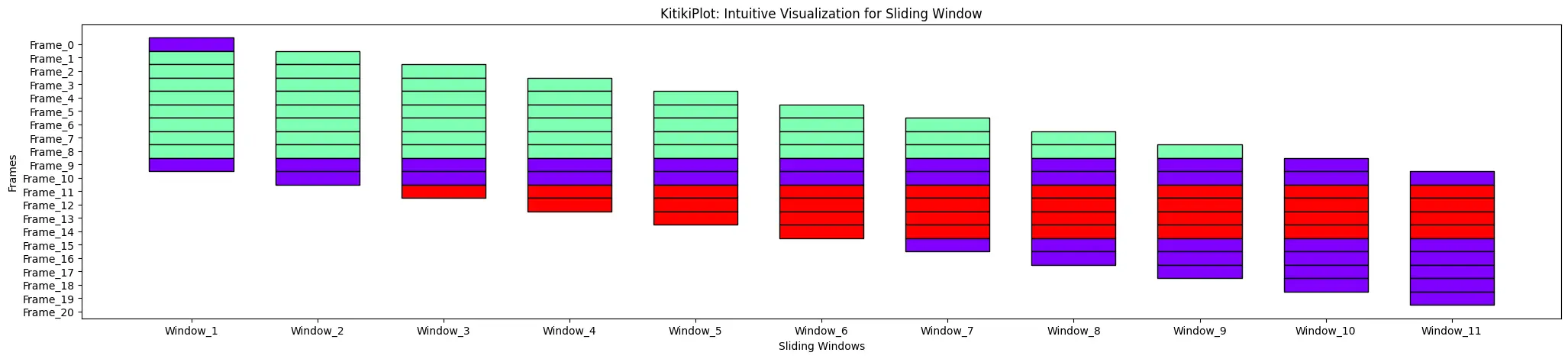
cell_height : float
- The peak of every cell within the grid.
- Default is 2.0.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( cell_height= 3 )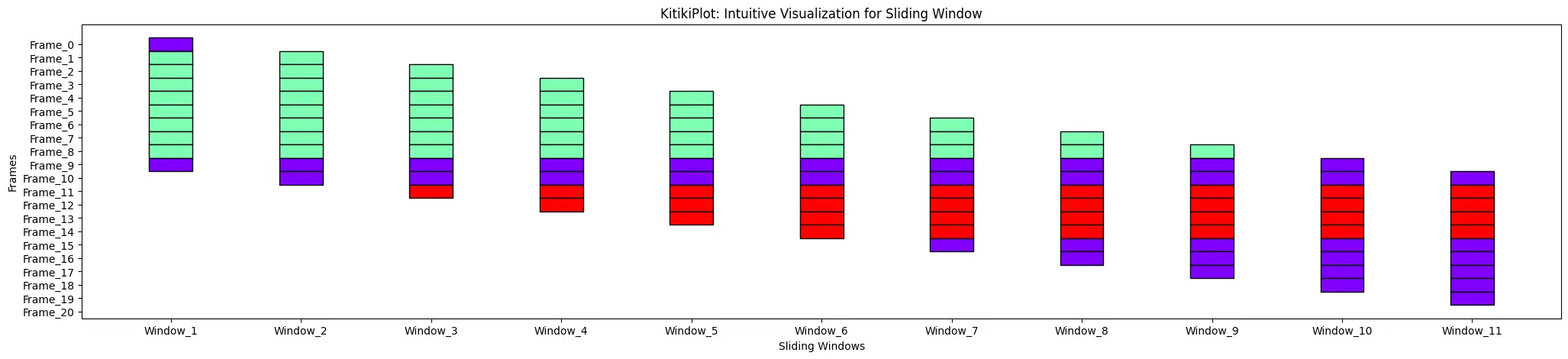
transpose : bool (optionally available)
- A flag indicating whether or not to transpose the KitikiPlot.
- Default is False.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( transpose= False )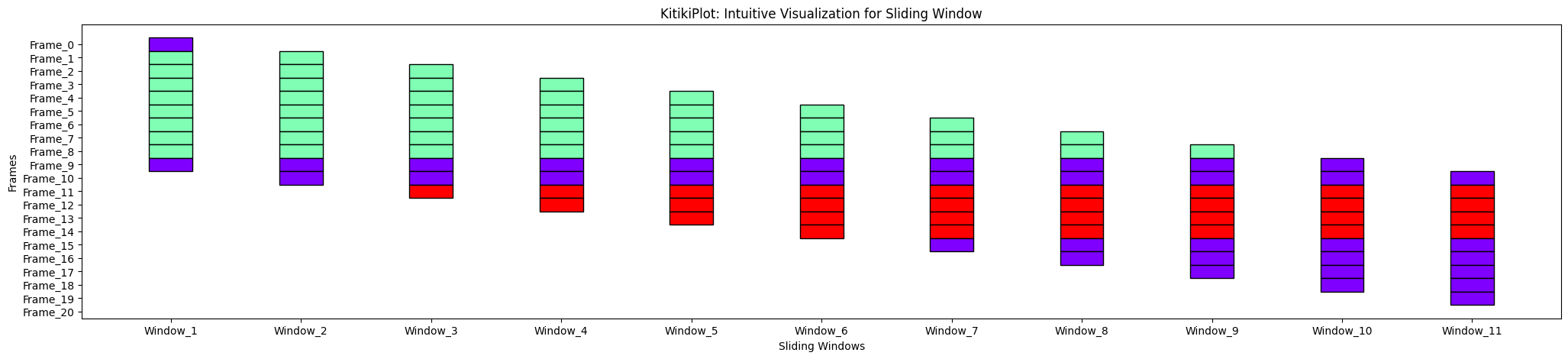
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot(
cell_width= 2,
transpose= True,
xtick_prefix= "Body",
ytick_prefix= "Window",
xlabel= "Frames",
ylabel= "Sliding_Windows" )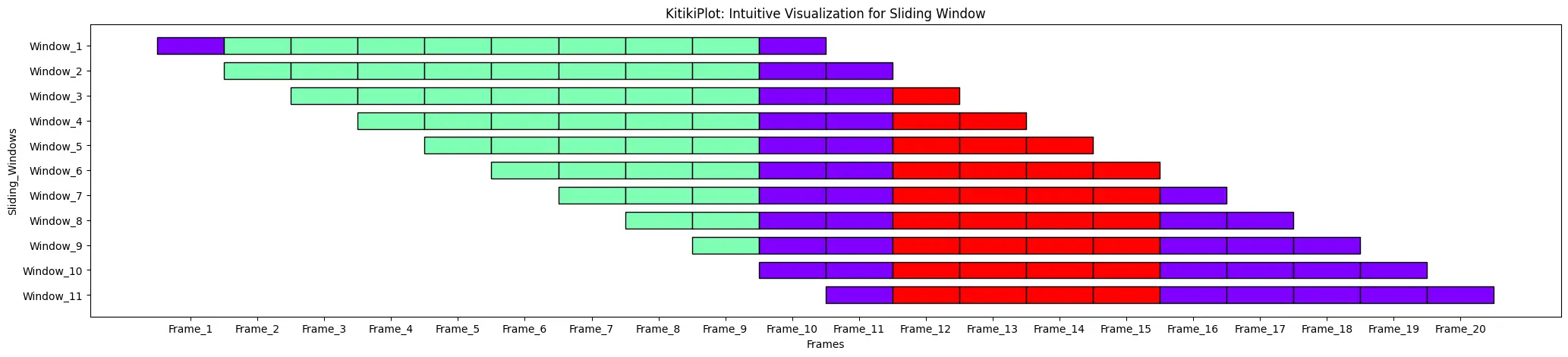
window_gap : float
- The hole between cells within the grid.
- Default is 1.0.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( window_gap= 3 )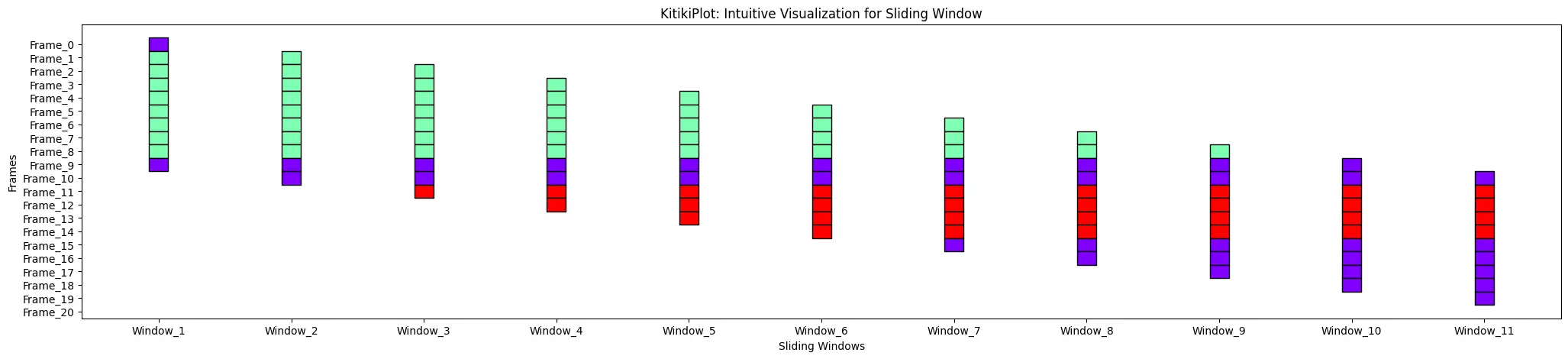
window_range : str or tuple (optionally available)
- The vary of home windows to show.
- Use “all” to point out all home windows or specify a tuple (start_index, end_index).
- Default is “all”.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( window_range= "all" )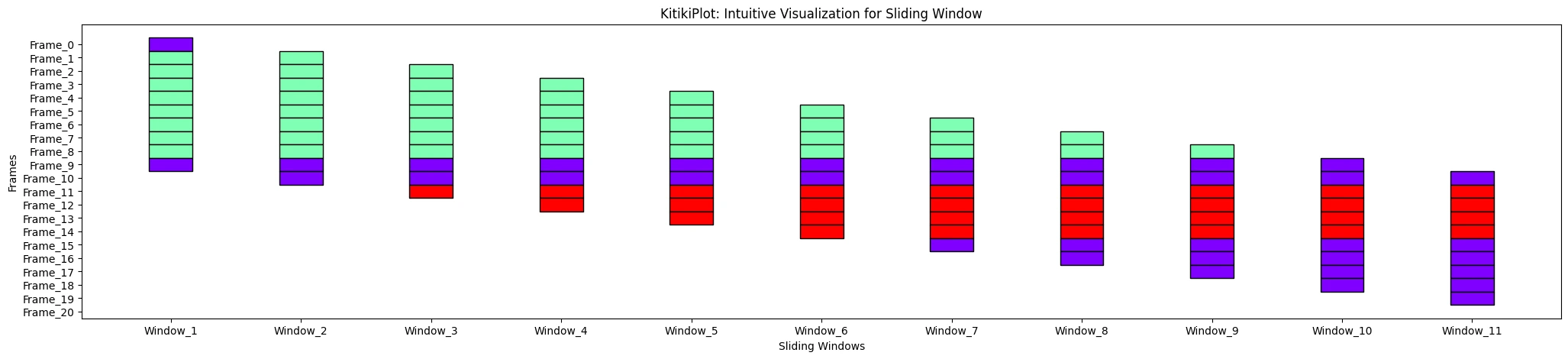
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( window_range= (3,8) )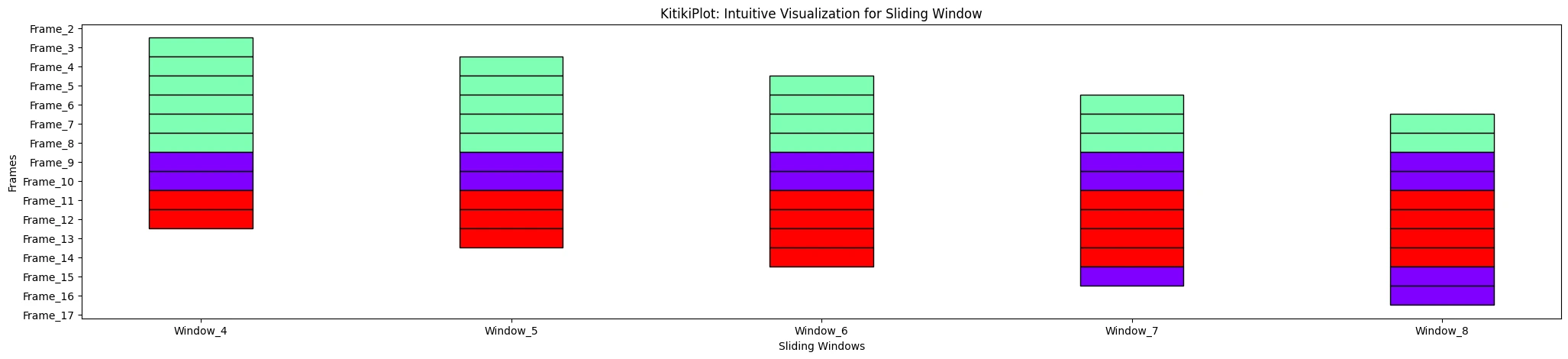
align : bool
- A flag indicating whether or not to shift consecutive bars vertically (if transpose= False), andhorizontally(if transpose= True) by stride worth.
- Default is True.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( align= True )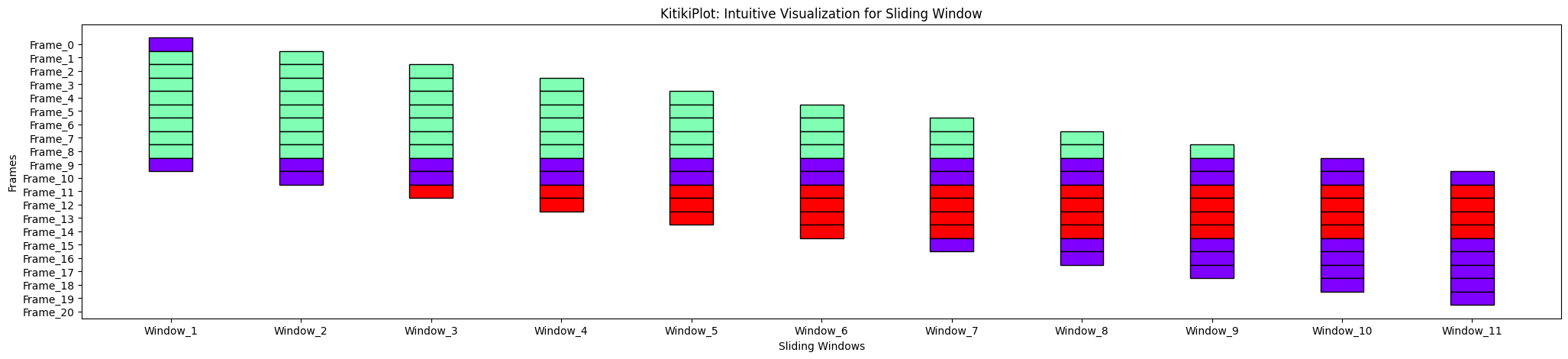
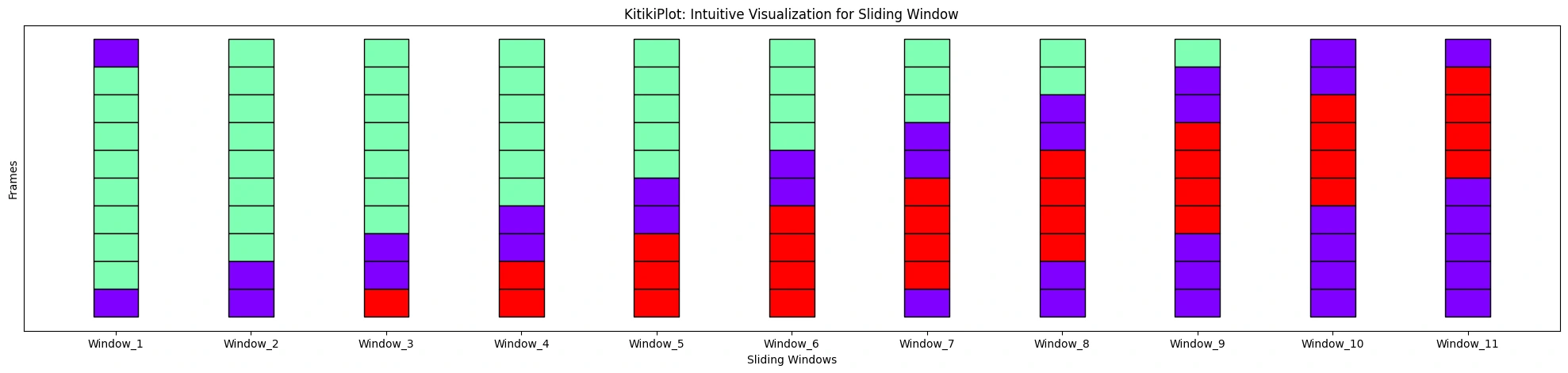
ktk.plot(
align= False,
display_yticks= False # Show no yticks
)
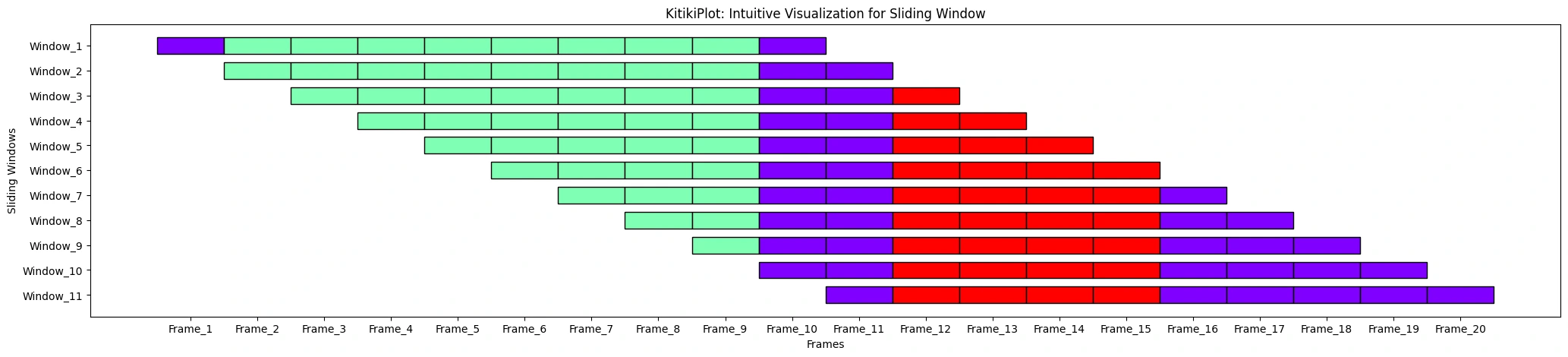
ktk.plot(
cell_width= 2,
align= True,
transpose= True,
xlabel= "Frames",
ylabel= "Sliding Home windows",
xtick_prefix= "Body",
ytick_prefix= "Window"
)
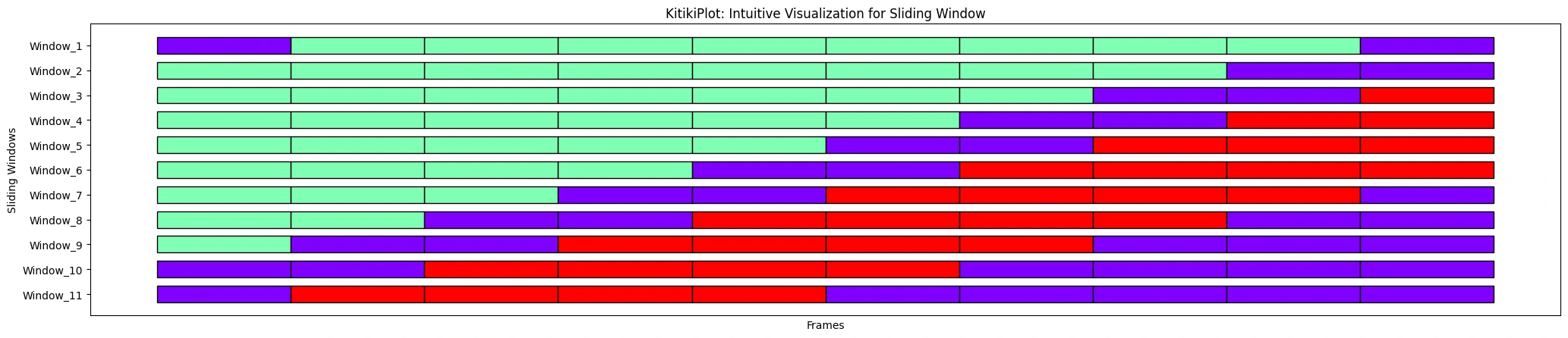
ktk.plot(
cell_width= 2,
align= False,
transpose= True,
xlabel= "Frames",
ylabel= "Sliding Home windows",
ytick_prefix= "Window",
display_xticks= False
)
cmap : str or dict
- If a string, it ought to be a colormap title to generate colours.
- If a dictionary, it ought to map distinctive values to particular colours.
- Default is ‘rainbow’.
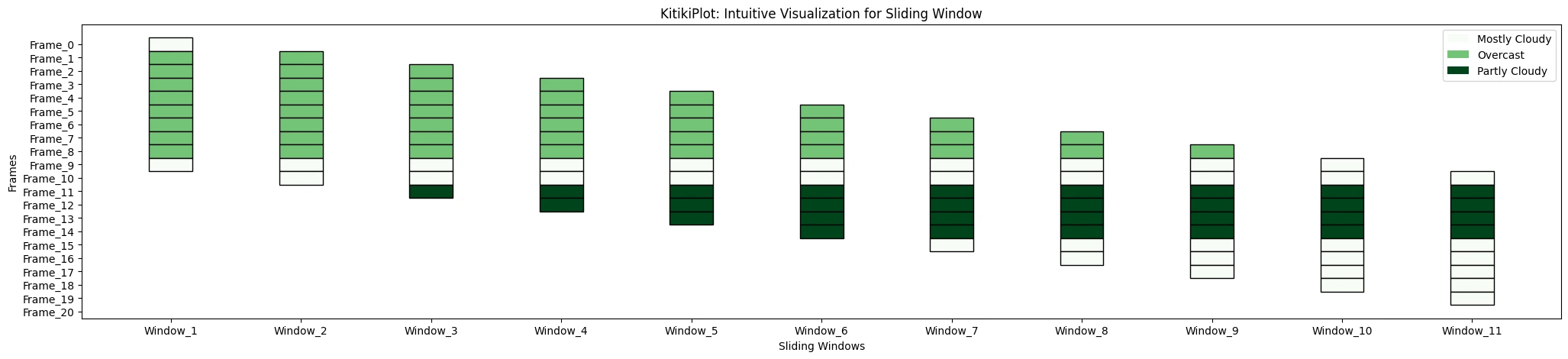
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot(
cmap= "Greens",
display_legend= True
)
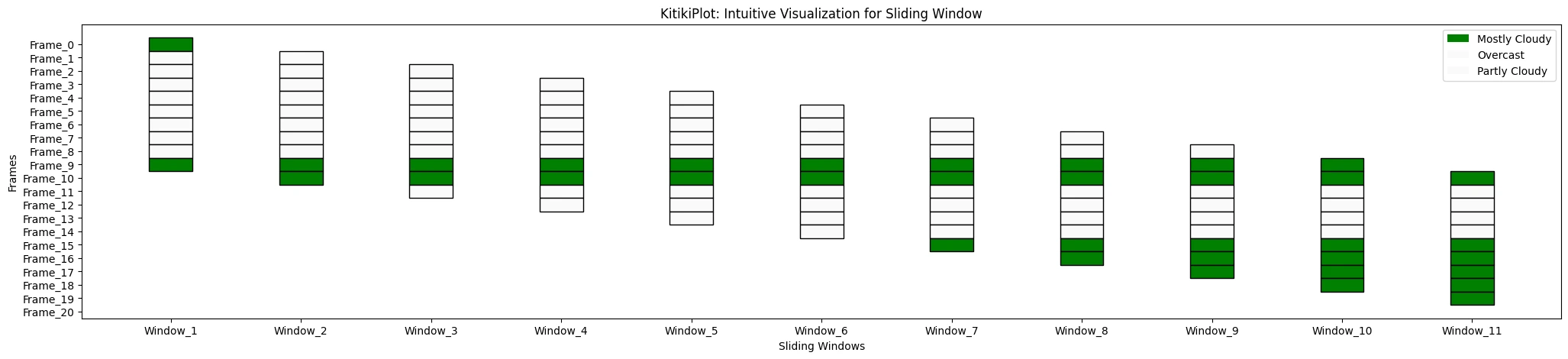
ktk.plot(
cmap= {"Principally Cloudy": "Inexperienced"},
display_legend= True
)
edge_color : str
- The colour to make use of for the sides of the rectangle.
- Default is ‘#000000’.
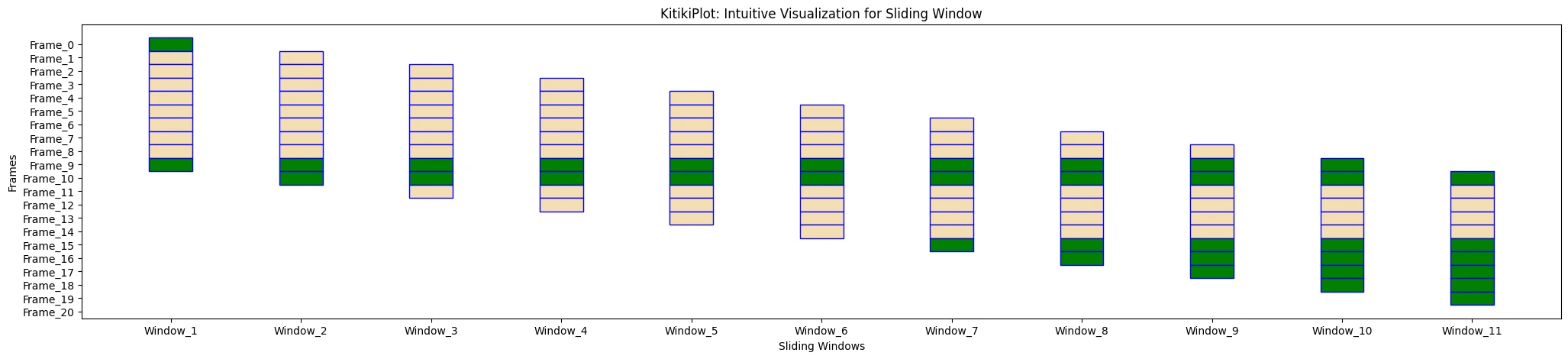
ktk.plot(
cmap= {"Principally Cloudy": "Inexperienced"},
fallback_color= "wheat",
edge_color= "blue",
)
fallback_color : str
- The colour to make use of as fallback if no particular colour is assigned.
- Default is ‘#FAFAFA’.
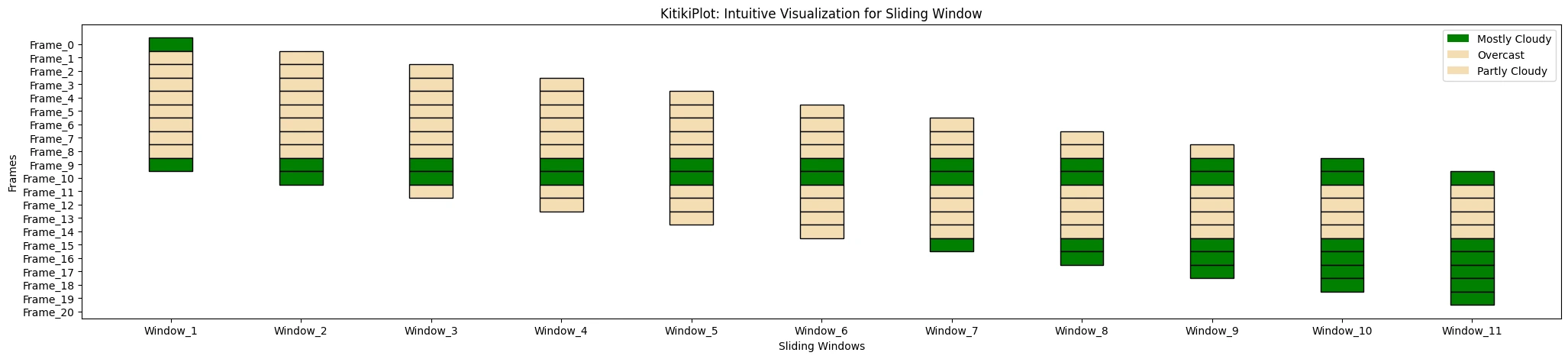
ktk.plot(
cmap= {"Principally Cloudy": "Inexperienced"},
fallback_color= "wheat",
display_legend= True
)
hmap : dict
- A dictionary mapping distinctive values to their corresponding hatch patterns.
- Default is ‘{}’.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot(
cmap= {"Principally Cloudy": "gray"},
fallback_color= "white",
hmap= ",
display_hatch= True
)
fallback_hatch : str
- The hatch sample to make use of as fallback if no particular hatch is assigned.
- Default is ‘” “‘ (string with single house).
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot(
cmap= {"Principally Cloudy": "gray"},
fallback_color= "white",
hmap= ",
fallback_hatch= "",
display_hatch= True
)
display_hatch : bool
- A flag indicating whether or not to show hatch patterns on cells.
- Default is False.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot(
cmap= {"Principally Cloudy": "#ffffff"},
fallback_color= "white",
display_hatch= True
)
xlabel : str (optionally available)
- Label for the x-axis.
- Default is “Sliding Home windows”.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( xlabel= "Commentary Window" )
ylabel : str (optionally available)
- Label for the y-axis.
- Default is “Frames”.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( ylabel= "Body ID" )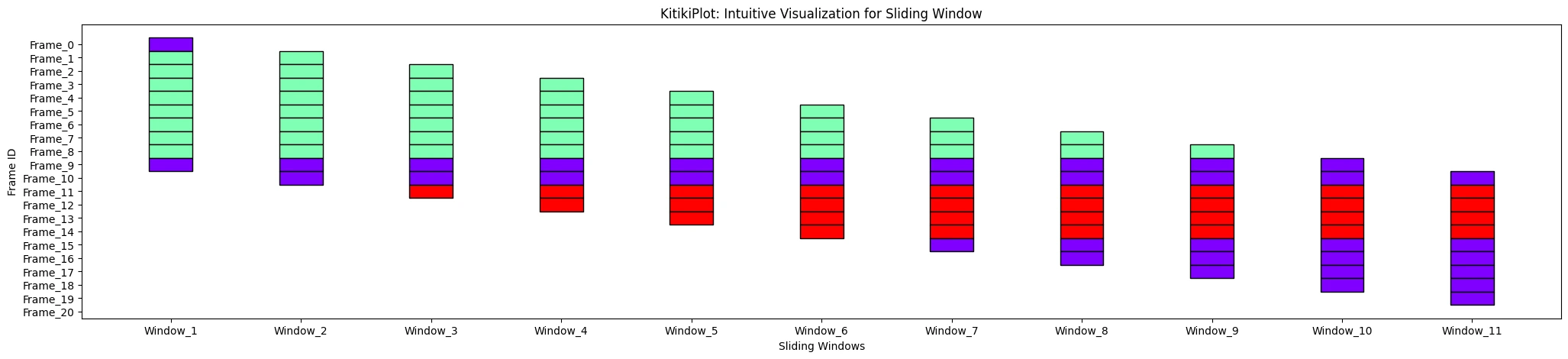
display_xticks : bool (optionally available)
- A flag indicating whether or not to show xticks.
- Default is True.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( display_xticks= False )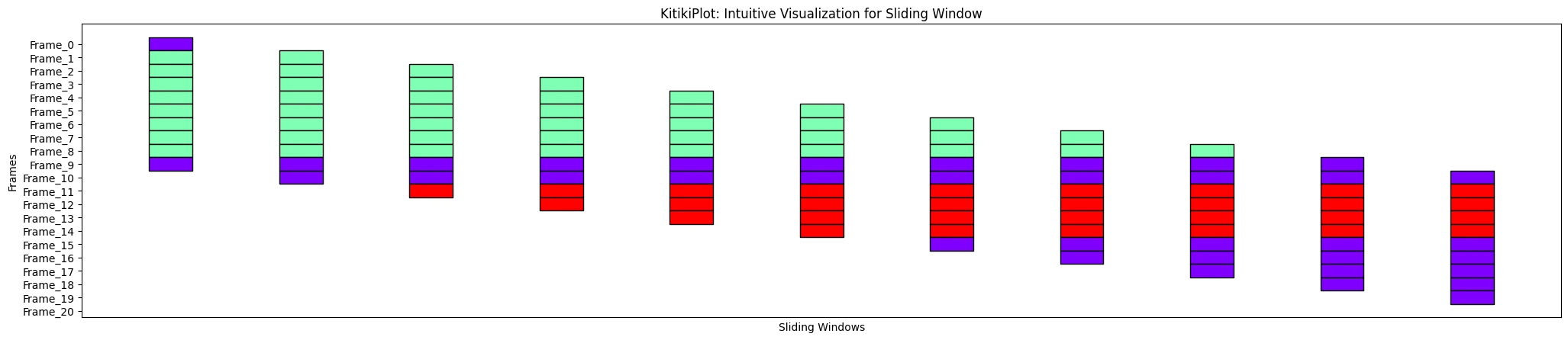
display_yticks : bool (optionally available)
- A flag indicating whether or not to show yticks
- Default is True
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( display_yticks= False )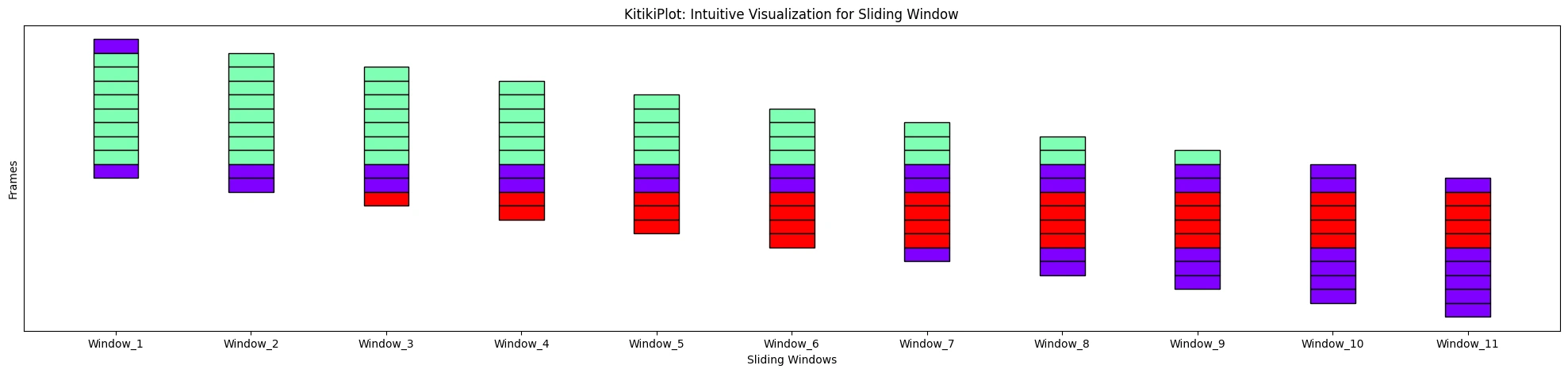
xtick_prefix : str (optionally available)
- Prefix for x-axis tick labels.
- Default is “Window”.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( xtick_prefix= "Commentary" )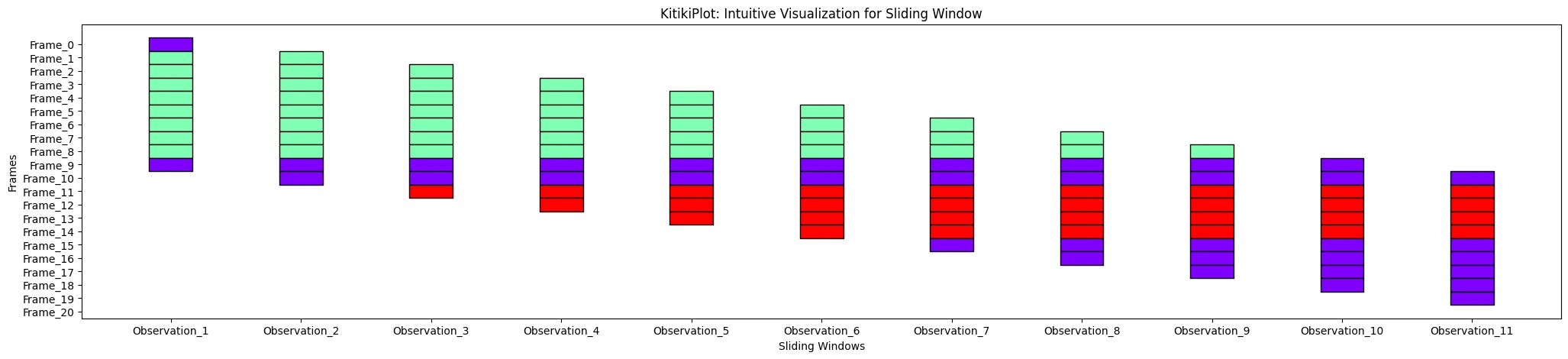
ytick_prefix : str (optionally available)
- Prefix for y-axis tick labels.
- Default is “Body”.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( ytick_prefix= "Time" )
xticks_values : checklist (optionally available)
- Listing containing the values for xticks
- Default is []
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
xtick_values= ['Start', "T1", "T2", "T3", "T4", "T5", "T6", "T7", "T8", "T9", "End"]
ktk.plot( xticks_values= xtick_values )
yticks_values : checklist (optionally available)
- Listing containing the values for yticks
- Default is []
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
yticks_values= [(str(i.hour)+" "+i.strftime("%p").lower()) for i in pd.to_datetime(df["Formatted Date"])]
ktk.plot( yticks_values= yticks_values )
xticks_rotation : int (optionally available)
- Rotation angle for x-axis tick labels.
- Default is 0.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( xticks_rotation= 45 )
yticks_rotation : int (optionally available)
- Rotation angle for y-axis tick labels.
- Default is 0.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( figsize= (20, 8), # Improve top of the plot for good visualization (right here)
yticks_rotation= 45
)
title : str (optionally available)
- The title of the plot.
- Default is “KitikiPlot: Intuitive Visualization for Sliding Window”.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( title= "KitikiPlot for Climate Dataset")
display_grid : bool (optionally available)
- A flag indicating whether or not to show grid on the plot.
- Default is False.
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot( display_grid= True )
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
cell_width= 2,
transpose= True,
xlabel= "Frames",
ylabel= "Sliding Home windows",
ytick_prefix= "Window",
display_xticks= False,
display_grid= True
)
display_legend : bool (optionally available)
- A flag indicating whether or not to show a legend on the plot.
- Default is False.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot( display_legend= True )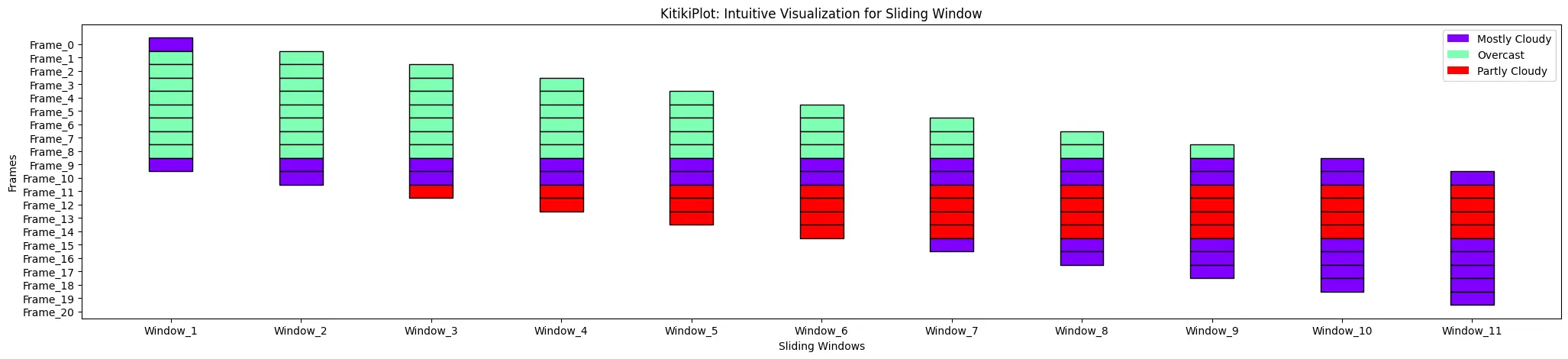
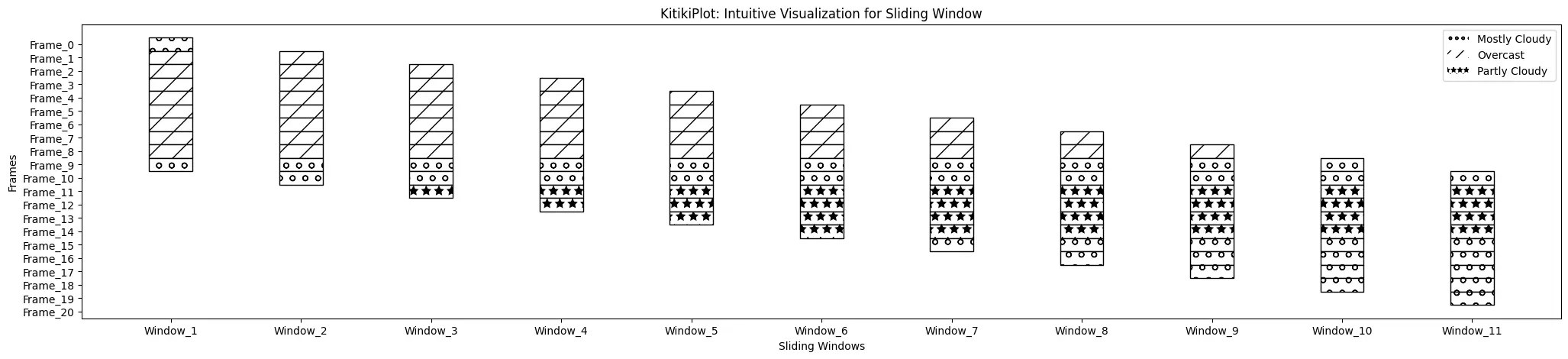
legend_hatch : bool (optionally available)
- A flag indicating whether or not to incorporate hatch patterns within the legend.
- Default is False.
ktk= KitikiPlot( information= df["Summary"].values.tolist() )
ktk.plot(
cmap= {"Principally Cloudy": "#ffffff"},
fallback_color= "white",
display_hatch= True,
display_legend= True,
legend_hatch= True
)
legend_kwargs : dict (optionally available)
- Further key phrase arguments handed to customise the legend.
- Default is {}.
Place legend exterior of the Plot
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
figsize= (15, 5),
display_legend= True,
legend_kwargs= {"bbox_to_anchor": (1, 1.0), "loc":"higher left"} )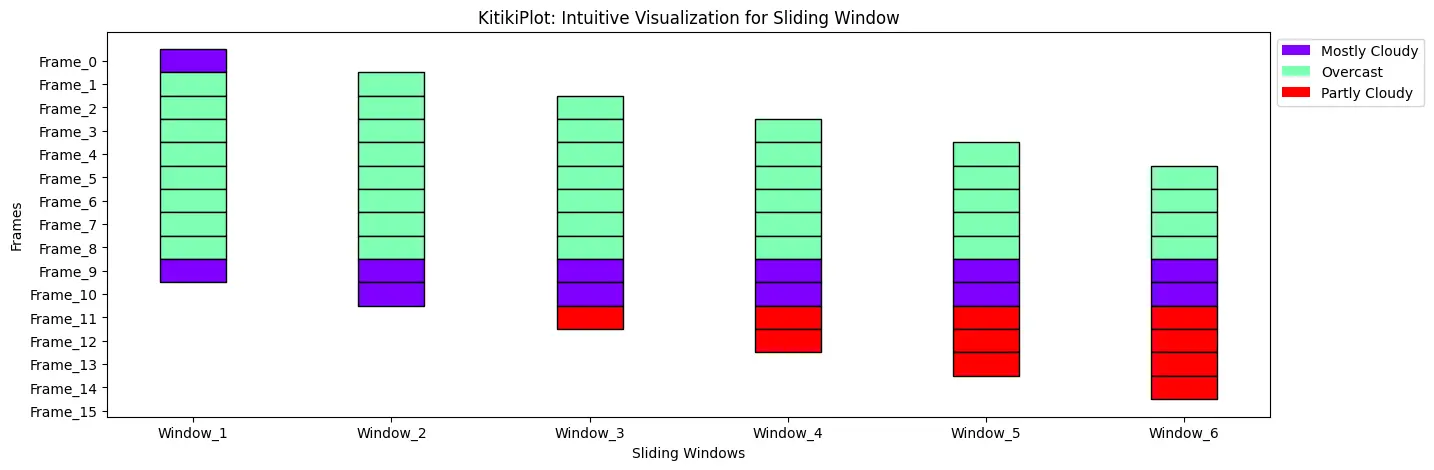
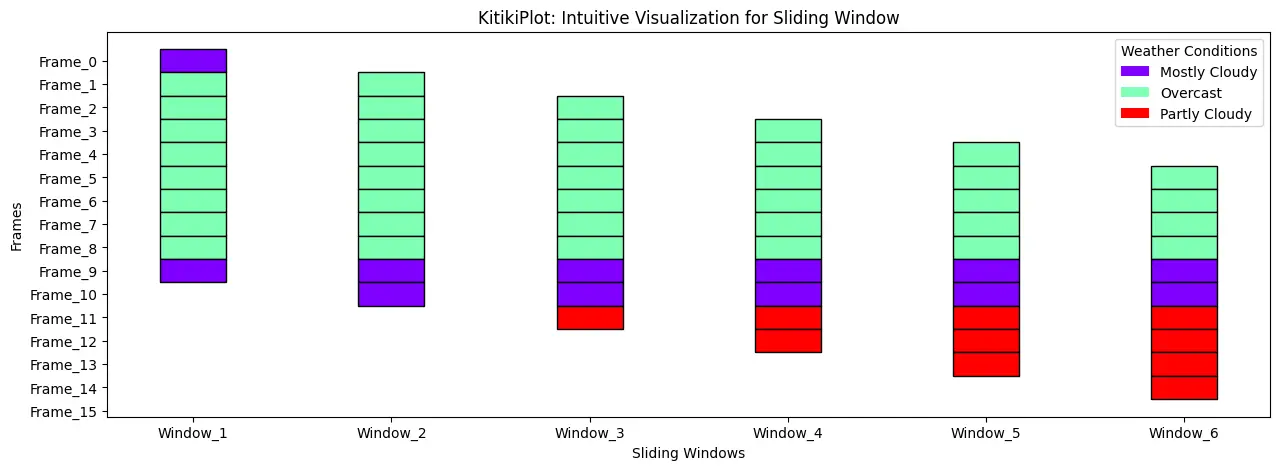
Set title for the legend————————————
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
figsize= (15, 5),
display_legend= True,
legend_kwargs= {"title": "Climate Situations"}
)
Change edgecolor of the legend
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
figsize= (15, 5),
display_legend= True,
legend_kwargs= {"edgecolor": "lime"}
)
kitiki_cell_kwargs : dict (optionally available)
- Further key phrase arguments handed to customise particular person cells.
- Default is {}.
Set the road model
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
figsize= (15, 5),
kitiki_cell_kwargs= {"linestyle": "--"} )
Modify the road width
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
figsize= (15, 5),
kitiki_cell_kwargs= {"linewidth": 3} )
Modify the alpha
ktk= KitikiPlot( information= df["Summary"].values.tolist()[:15] )
ktk.plot(
figsize= (15, 5),
kitiki_cell_kwargs= {"alpha": 0.4} )
Actual-World Functions of KitikiPlot
KitikiPlot excels in numerous fields the place information visualization is essential to understanding complicated patterns and tendencies. From genomics and environmental monitoring to finance and predictive modeling, KitikiPlot empowers customers to rework uncooked information into clear, actionable insights. Whether or not you’re analyzing giant datasets, monitoring air high quality over time, or visualizing tendencies in inventory costs, KitikiPlot presents the flexibleness and customization wanted to satisfy the distinctive calls for of varied industries.
Genomics
- KitikiPlot permits clear visualization of gene sequences, serving to researchers establish patterns and motifs.
- It facilitates the evaluation of structural variations in genomes, important for understanding genetic problems.
- By offering visible representations, it aids in deciphering complicated genomic information, supporting developments in personalised medication.
Dataset URL: https://archive.ics.uci.edu/dataset/69/molecular+biology+splice+junction+gene+sequences
# Import essential libraries
from kitikiplot import KitikiPlot
import pandas as pd
# Load the dataset
df= pd.read_csv( "datasets/molecular+biology+splice+junction+gene+sequences/splice.information", header= None )
# Rename the columns
df.columns= ["Label", "Instance_Name", "Nucleotide_Sequence"]
# Choose 3 gene sequences randomly
df= df.pattern(3, random_state= 1)
# Take away the white areas from the "Nucleotide_Sequence"
df["Nucleotide_Sequence"]= df["Nucleotide_Sequence"].str.strip()
df
index= 0
ktk= KitikiPlot( information= [i for i in df.iloc[index, 2]], stride= 1, window_length= len(df.iloc[index, 2]) )
ktk.plot(
figsize= (20, 0.5),
cell_width= 2,
cmap= {'A': '#007FFF', 'T': "#fffc00", "G": "#00ff00", "C": "#960018"},
transpose= True,
xlabel= "Nucleotides",
ylabel= "Sequence",
display_yticks= False,
xtick_prefix= "Nucleotide",
xticks_rotation= 90,
title= "Genome Visualization: "+df.iloc[index, 1].strip()+", Label : "+df.iloc[index,0].strip(),
display_legend= True,
legend_kwargs= {"bbox_to_anchor": (1.01, 1), "loc":'higher left', "borderaxespad": 0.})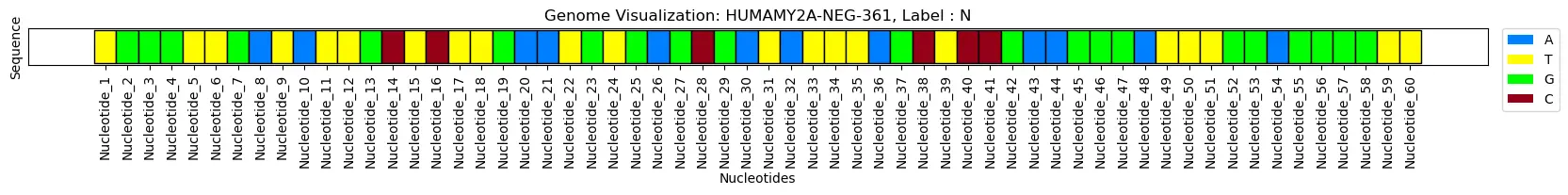
Climate Forecasting
- The library can successfully signify temporal climate information, reminiscent of temperature and humidity, over sequential time home windows to establish tendencies.
- This visualization aids in detecting patterns and fluctuations in climate situations, enhancing the accuracy of forecasts.
- Moreover, it helps the evaluation of historic information, permitting for higher predictions and knowledgeable decision-making relating to weather-related actions.
Dataset URL: https://www.kaggle.com/datasets/muthuj7/weather-dataset
# Import essential libraries
from kitikiplot import KitikiPlot
import pandas as pd
# Learn csv
df= pd.read_csv( "datasets/weatherHistory/weatherHistory.csv")
print("Form: ", df.form)
# Choose a subset of knowledge for visualization
df= df.iloc[45:65, :]
print("Form: ", df.form)
df.head(3)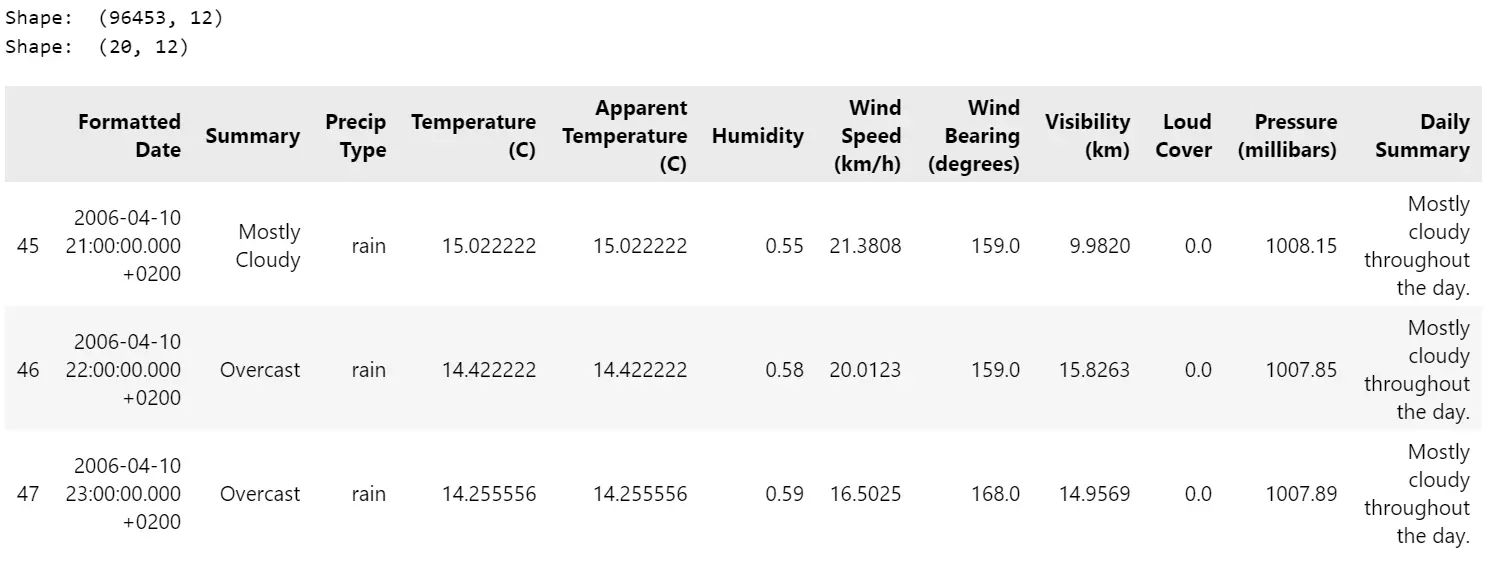
index= 0
weather_data= ['Mostly Cloudy', 'Overcast', 'Overcast', 'Overcast', 'Overcast', 'Overcast','Overcast', 'Overcast',
'Overcast', 'Mostly Cloudy', 'Mostly Cloudy', 'Partly Cloudy', 'Partly Cloudy', 'Partly Cloudy',
'Partly Cloudy', 'Mostly Cloudy', 'Mostly Cloudy', 'Mostly Cloudy', 'Mostly Cloudy', 'Mostly Cloudy']
time_period= ['21 pm', '22 pm', '23 pm', '0 am', '1 am', '2 am', '3 am', '4 am', '5 am', '6 am', '7 am', '8 am',
'9 am', '10 am', '11 am', '12 pm', '13 pm', '14 pm', '15 pm', '16 pm']
ktk= KitikiPlot( information= weather_data, stride= 1, window_length= 10 )
ktk.plot(
figsize= (20, 5),
cell_width= 2,
transpose= False,
xlabel= "Window",
ylabel= "Time",
yticks_values= time_period,
xticks_rotation= 90,
cmap= {"Principally Cloudy": "brown", "Partly Cloudy": "#a9cbe0","Overcast": "#fdbf6f"},
legend_kwargs= {"bbox_to_anchor": (1.01, 1), "loc":'higher left', "borderaxespad": 0.},
display_legend= True,
title= "Climate Sample on 10-04-2006")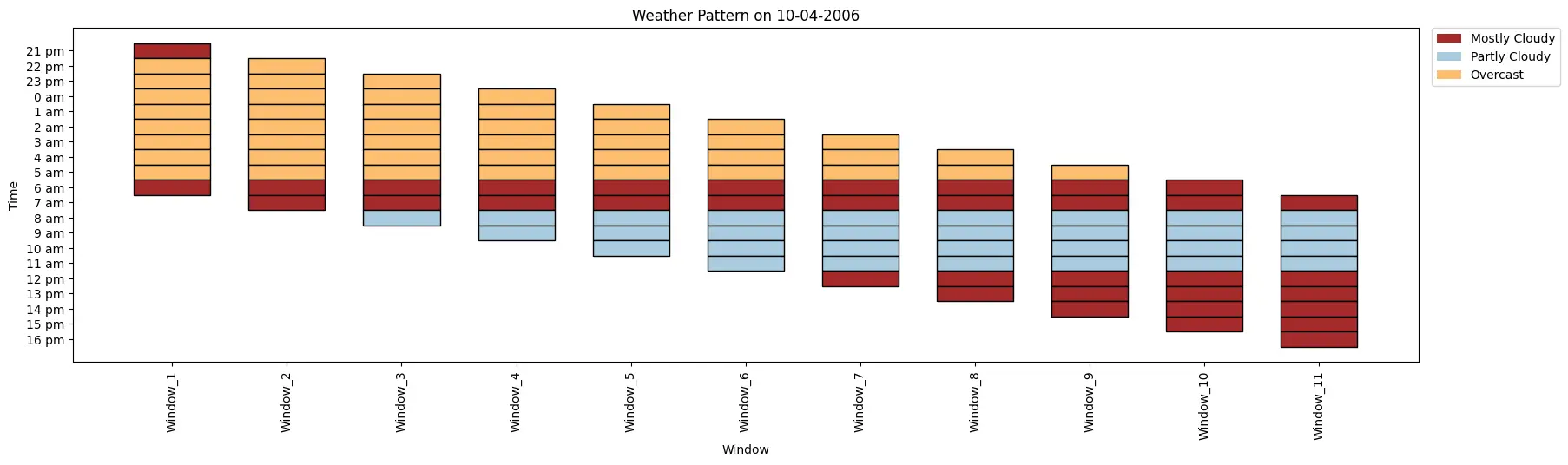
Air High quality Monitoring
- Customers can analyze pollutant ranges over time with KitikiPlot to detect variations and correlations in environmental information.
- This functionality permits for the identification of tendencies in air high quality, facilitating a deeper understanding of how completely different pollution work together and fluctuate as a result of varied elements.
- Moreover, it helps the exploration of temporal relationships between air high quality indices and particular pollution, enhancing the effectiveness of air high quality monitoring efforts.
Dataset URL: https://archive.ics.uci.edu/dataset/360/air+high quality
from kitikiplot import KitikiPlot
import pandas as pd
# Learn excel
df= pd.read_excel( "datasets/air+high quality/AirQualityUCI.xlsx" )
# Extract information from at some point (2004-11-01)
df= df[ df['Date']== "2004-11-01" ]
print("Form : ", df.form)
df.head( 3 )
# Convert float to int
df["CO(GT)"]= df["CO(GT)"].astype(int)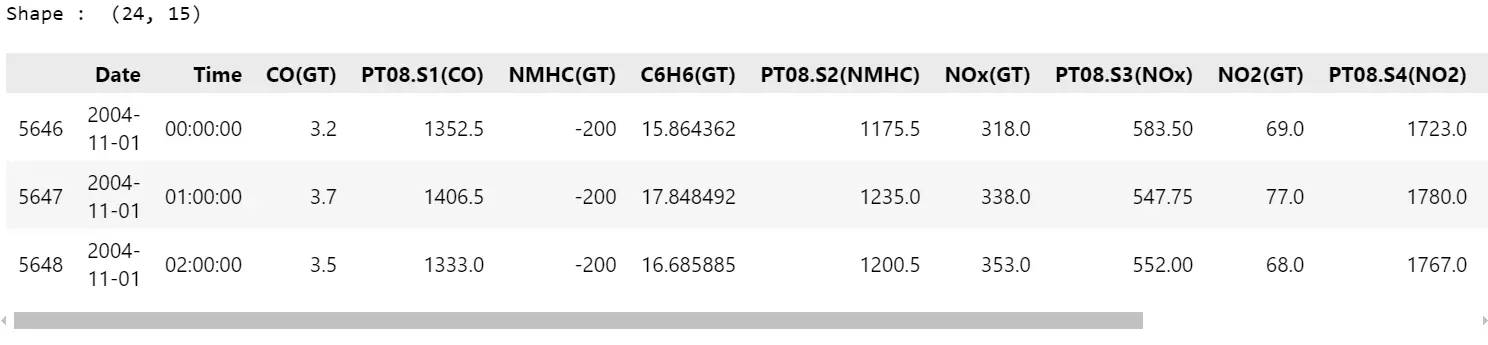
CO_values= [3, 3, 3, 3, -200, 2, 1, 1, 1, 1, 2, 3, 4, 4, 3, 3, 3, 2]
time_period= ['0 am', '1 am', '2 am', '3 am', '4 am', '5 am', '6 am', '7 am', '8 am', '9 am', '10 am',
'11 am', '12 pm', '13 pm', '14 pm', '15 pm', '16 pm', '17 pm']
ktk= KitikiPlot( information= CO_values )
ktk.plot(
figsize= (20, 5),
cell_width= 2,
cmap= {-200: "cornflowerblue", 1: "#ffeaea", 2: "#feb9b9", 3: "#ff7070", 4: "#b00"},
transpose= True,
xlabel= "Time",
ylabel= "Sliding Home windows of CO(GT) values (in mg/m^3)",
display_xticks= True,
xticks_values= time_period,
ytick_prefix= "Window",
xticks_rotation= 90,
display_legend= True,
title= "CO(GT) Development in Air",
legend_kwargs= {"bbox_to_anchor": (1.01, 1), "loc":'higher left', "borderaxespad": 0.})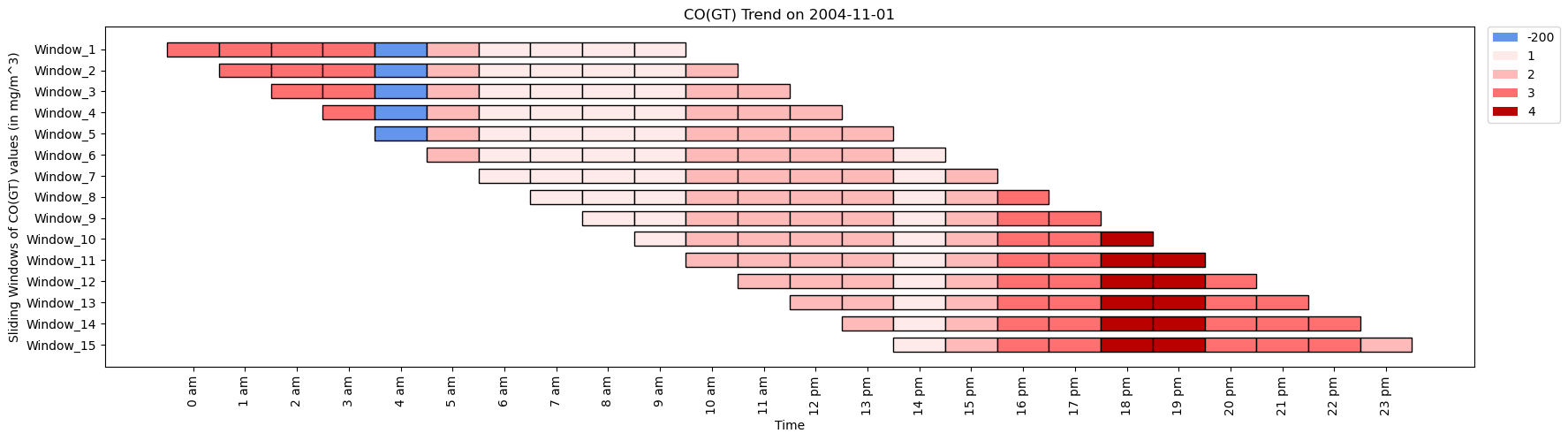
Conclusion
KitikiPlot simplifies the visualization of sequential and time-series categorical sliding window information, making complicated patterns extra interpretable. Its versatility spans varied purposes, together with genomics, climate evaluation, and air high quality monitoring, highlighting its broad utility in each analysis and trade. With a deal with readability and usefulness, KitikiPlot enhances the extraction of actionable insights from categorical information. As an open-source library, it empowers information scientists and researchers to successfully deal with numerous challenges.
Key Takeaways
- KitikiPlot is a flexible Python library designed for exact and user-friendly sliding window information visualizations.
- Its customizable parameters permit customers to create significant and interpretable visualizations tailor-made to their datasets.
- The library helps a variety of real-world purposes throughout varied analysis and trade domains.
- As an open-source device, KitikiPlot ensures accessibility for information science practitioners and researchers alike.
- Clear and insightful visualizations facilitate the identification of tendencies in sequential categorical information.
Sources
Quotation
@software program{ KitikiPlot_2024
writer = {Boddu Sri Pavan and Boddu Swathi Sree},
title = {{KitikiPlot: A Python library to visualise categorical sliding window information}},
12 months = {2024},
model = {0.1.2},
url = {url{https://github.com/BodduSriPavan-111/kitikiplot},
doi = {10.5281/zenodo.14293030}
howpublished = {url{https://github.com/BodduSriPavan-111/kitikiplot}}
}Ceaselessly Requested Questions
A. KitikiPlot makes a speciality of visualizing sequential and time-series categorical information utilizing a sliding window strategy.
A. Whereas primarily supposed for categorical information, KitikiPlot might be tailored for different information sorts by means of artistic preprocessing strategies, reminiscent of discretization, and so forth,.
A. Sure, KitikiPlot integrates seamlessly with common libraries like Pandas and Matplotlib for efficient preprocessing and enhanced visualization.
The media proven on this article is just not owned by Analytics Vidhya and is used on the Writer’s discretion.